About
I'm a data scientist with a strong background in computer science and machine learning, and with 5+ years of relevant experience on both instruction and implementation of machine learning and software workflows.

Data Scientist
I work on large datasets and develop tools using machine learning and statistical methods to uncover patterns and to generate insights from data.
Computer Scientist
I have deep understanding of computer science fundamentals: programming, data structures, linear algebra and calculus, complexity and computability, databases, networks, numerical analysis, and machine learning.
Software Engineer
I have years of experience in software engineering (full-stack), with exposure to a wide array of technologies and frameworks, in both agile and waterfall environments.
PhD Candidate
I am currently working towards a PhD degree at the University of Oxford. I am analyzing single cell variations and cell-cell interactions in 3D histology using a combination of convolutional neural networks and statistical analyses.
Skills & Technologies
Data Science
Pytorch, Keras/Tensorflow CNN, RNN, VAE, GAN Scikit-learn Numpy Matplotlib, Tableau MLflow SQL Statistical sampling and testsSoftware Development
HTML CSS JS AWS Git Swift Django LAMP CI/CDProgramming
Python C/C++ Java Ruby PHP BashCV
Education
Doctor of Philosophy (Engineering Science, Biomedical Imaging)
2018 - 2023 (Expected)
University of Oxford, United Kingdom
Ph.D. research in engineering science (biomedical imaging) under the supervision of Prof. Jens Rittscher
THESIS: Learn representations of cell morphology using machine learning techniques to analyze time-series and 3D imaging microscopy and histology data and build tools (using Python, pytorch, numpy, scikit-learn) to support biological and medical hypotheses, particularly on DNA damage and immune response in tumor microenvironments.
Master of Science (Computer Science)
2013 - 2017
University of the Philippines-Diliman, Philippines
Bachelor of Science (Computer Science)
2010 - 2014
University of the Philippines-Diliman, Philippines
Professional Experience
Research Assistant
2021 - Present
University of Oxford, United Kingdom
- Built deep learning tools and performed statistical analysis to process large datasets (40+ terabytes of 3D histology imaging data) to effectively differentiate between control and treated tissue samples.
- Facilitated deployment and integration of machine learning tools and Python scripts in the National Institutes of Health for experimental medicine and biological research.
- Optimized and calibrated experimental setups using data analytics on imaging parameters and image data resolution.
- Visualized data and communicated the analysis results and potential clinical impact to both experts and non-experts in the biomedical imaging field at an international conference.
Computer Science Instructor
2013 - 2018
University of the Philippines-Diliman, Philippines
- Instructed 300+ students on computer science fundamentals (Computer Programming, Artificial Intelligence, Assembly Language Programming, Computer Organization, Data Structures, Programming Languages).
- Assisted in the revision of the computer science curriculum, in particular the artificial intelligence and machine learning course, to bring it up-to-date to newer technologies and industry requirements.
Software Developer
2017 - 2018
University of the Philippines-Diliman, Philippines
- Performed data modeling from gathered university requirements for staff management, developed and maintained existing core modules (PHP, HTML, CSS, JS) and databases (PostgreSQL) for university class registration.
Software Engineer
2016 - 2018
SUPost, USA
- Maintained code (bash, PHP, Python, and Ruby) and mySQL database for SUPost e-commerce platform with automated deployment and health-checks of server instances in cloud computing platforms.
- Designed a cloud-based NoSQL database to speed up queries and display of new advertisements by 15%.
- Designed and developed an iOS app using Swift to complement the web e-commerce application.
Research Assistant
2015 - 2017
University of the Philippines-Manila, Philippines
- Built machine learning tools using Tensorflow to detect key regions in microscopy images, integrated tools with existing web (Django) and Android platforms for remote diagnosis.
- Migrated and deployed web application and machine learning tools to AWS.
Portfolio
- All
- Research
- Software
- Poster
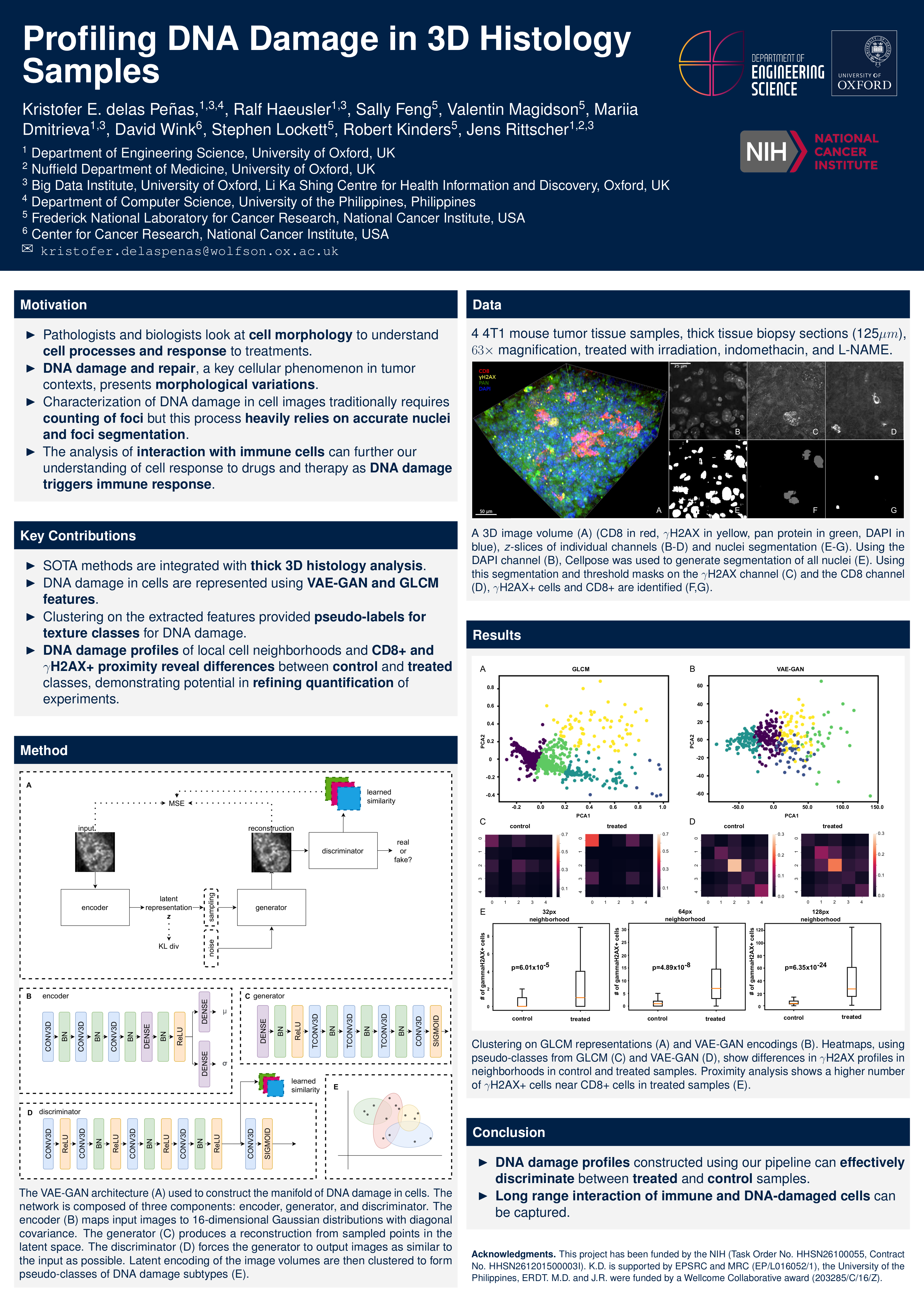
Kristofer E. delas Penas, Ralf Haeusler, Sally Feng, Valentin Magidson, Mariia Dmitrieva, David Wink, Stephen Lockett, Robert Kinders, Jens Rittscher Presented at MOVI workshop at MICCAI 2022
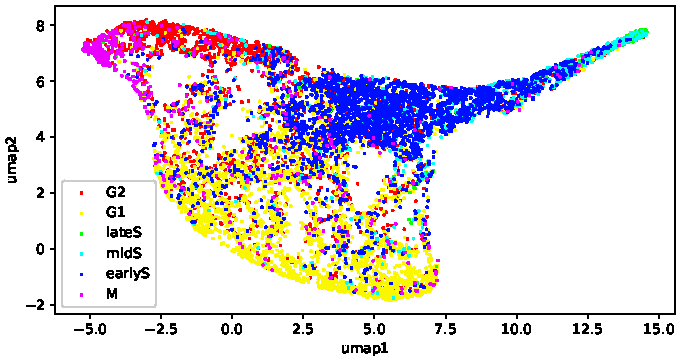
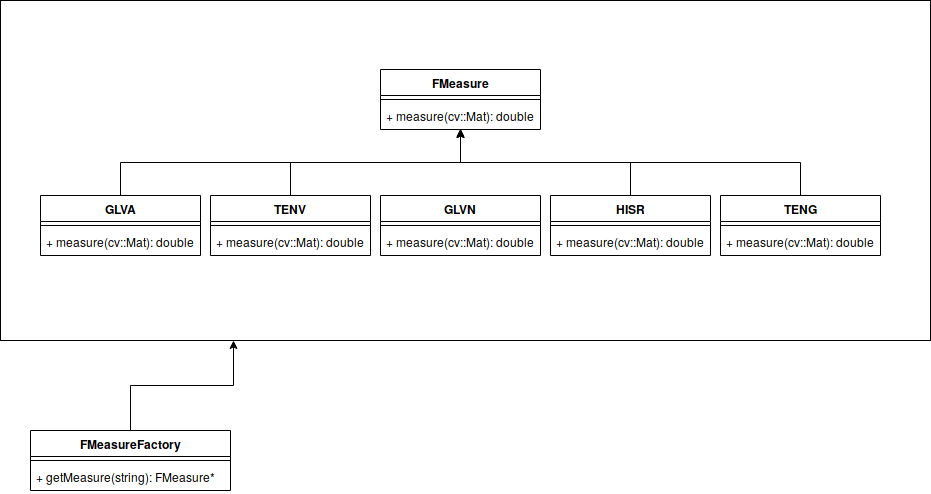
Kristofer delas Peñas, Mariia Dmitrieva, Dominic Waithe, Jens Rittscher Manuscript submitted
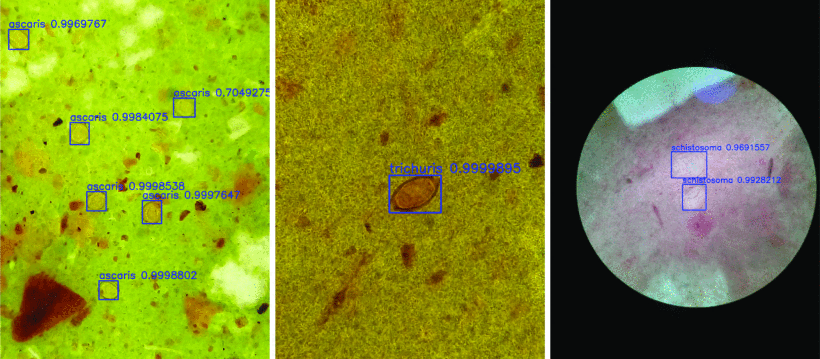
Kristofer delas Peñas, Elena Villacorte, Pilarita Rivera, Prospero Naval 2020 IEEE Region 10 Conference (TENCON), Osaka, Japan
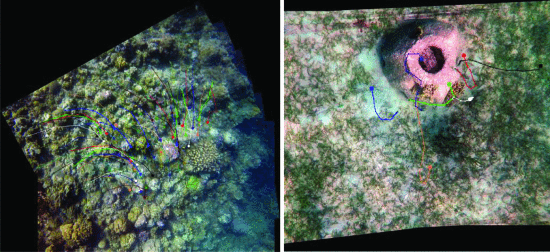
Riza Rae Pineda, Kristofer delas Peñas, Dana Manogan 2020 IEEE Region 10 Conference (TENCON), Osaka, Japan
Kristofer Delas Penas, Mariia Dmitrieva, Joël Lefebvre, Helen Zenner, Edward Allgeyer, Martin Booth, Daniel St. Johnston, Jens Rittscher 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Coralville, IA, USA
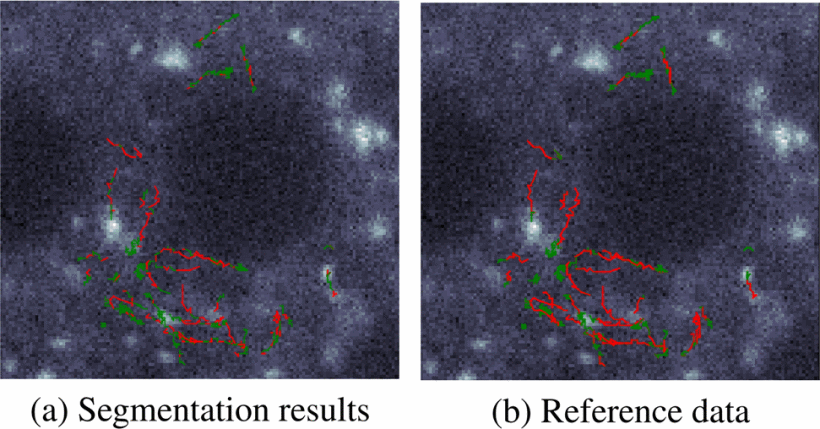
Mariia Dmitrieva, Joël Lefebvre, Kristofer Delas Penas, Helen Zenner, Jennifer Richens, Daniel St. Johnston, Jens Rittscher 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Coralville, IA, USA
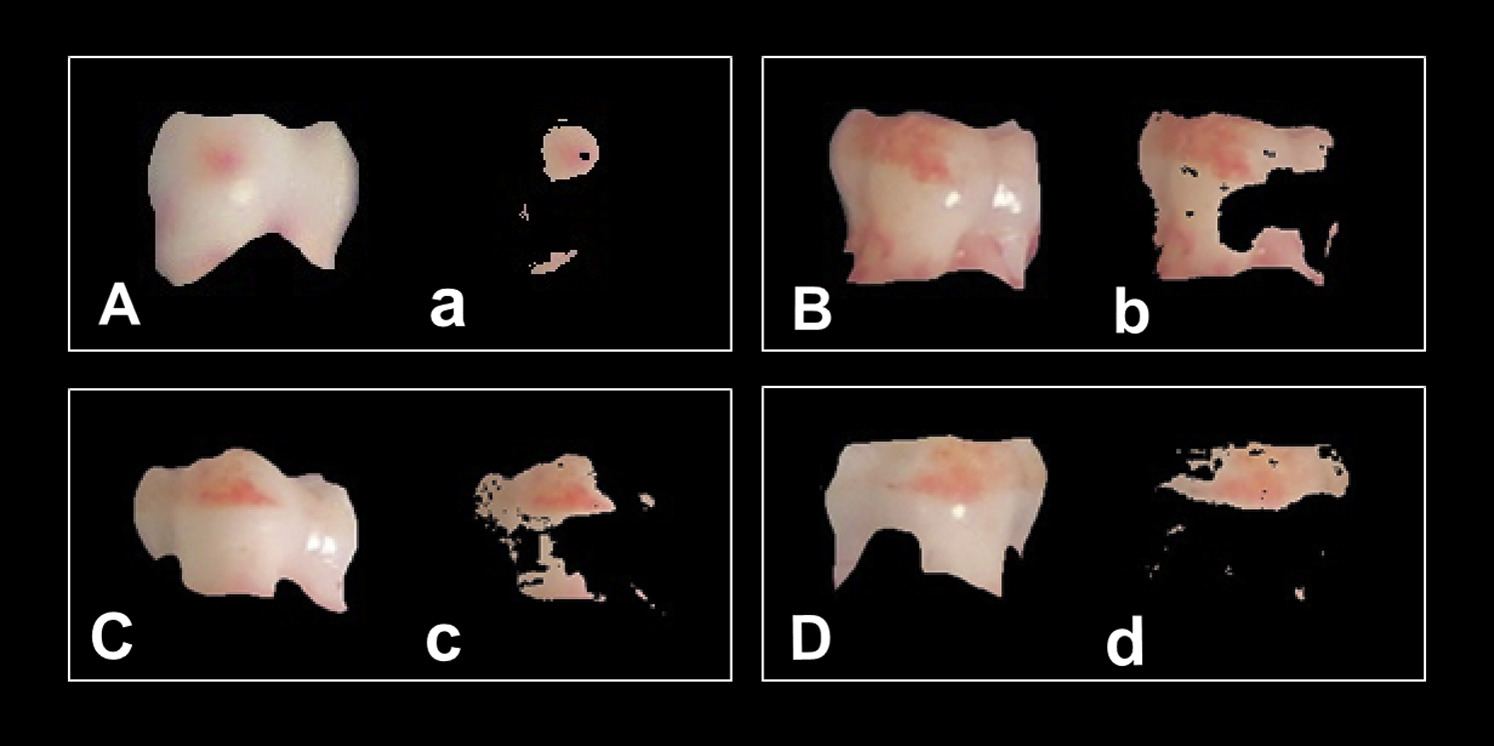
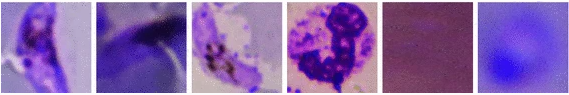
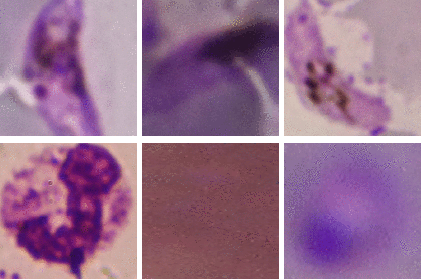
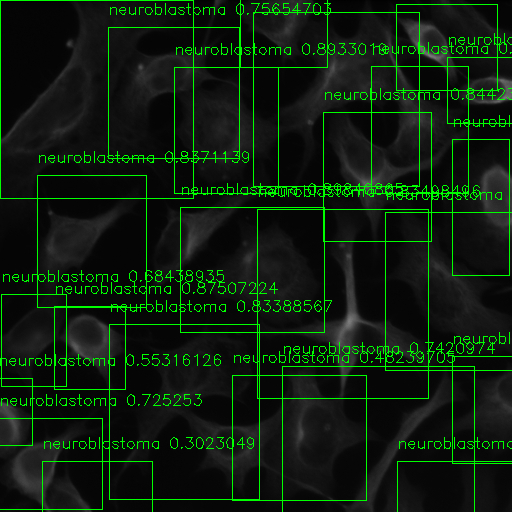
Kristofer delas Penas, Dominic Waithe